Matthew W. Kelley, Ph.D., Chief
Research Statement
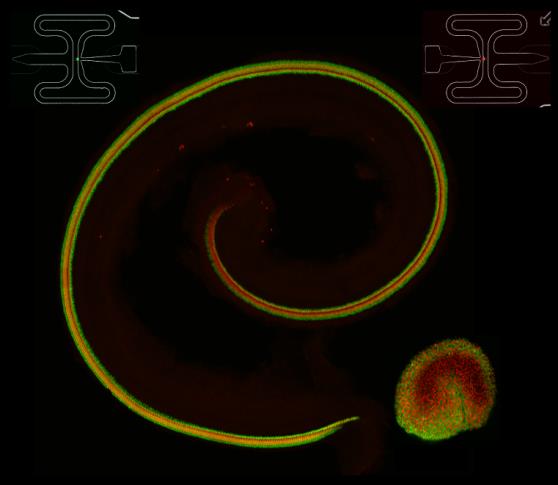
Cochlea and utricle from a transgenic animal in which all hair cells express td-Tomato and all supporting cells express green fluorescent protein.
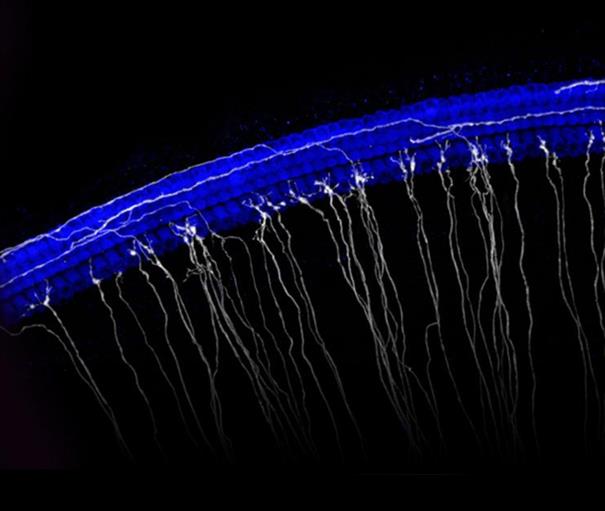
Image of the sensory epithelium of the cochlea. Hair cells are in blue and a subset of spiral ganglion neurons are in white.
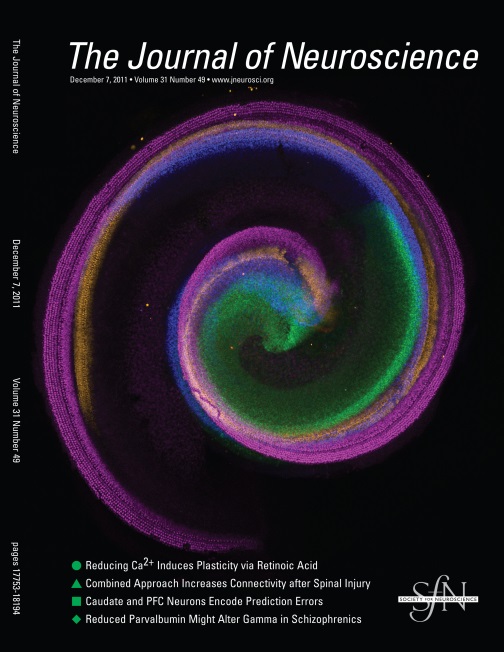
Cover image illustrating changes in the distribution of Sox2-positive cells during cochlear development. Different ages are depicted in different colors.
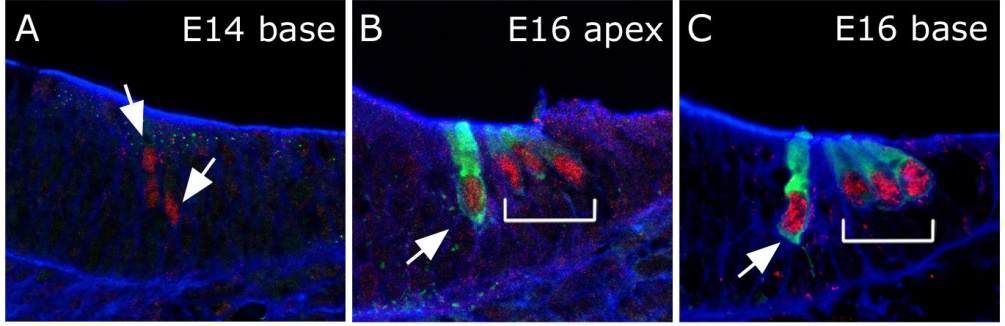
Cross sections through the cochlear duct illustrating expression of the transcription factor Atoh1 in red and the motor protein Myosin 7a in green.
The overall goals of the Section on Developmental Neuroscience (Laboratory of Cochlear Development) are to identify the molecular and cellular factors that play a role in the development of the different structures within the cochlea and inner ear, including the auditory and vestibular sensory epithelia and the spiral ganglion, which acts as the first relay between the inner ear and the brain. The auditory sensory epithelia, also called the organ of Corti, is composed of at least 6 distinct cell types arranged in highly conserved mosaic. The generation of a specific number of each cell type and the arrangement of these cell types into a regular pattern are essential for the normal perception of sound; however, our understanding of the factors that play a role in the development of this structure, and each of the unique cell types, is extremely limited.
Current research in the laboratory combines single cell isolation with RNA-Seq technology to generate profiles for unique cell types within auditory and vestibular epithelia and spiral ganglion neurons. Previous results have demonstrated that the number of cells that will develop as sensory hair cells is regulated through inhibitory interactions between neighboring cells. These results suggest that the various cell fates within the cochlea may be arranged in a hierarchy, and that as the number of cells that become specified to develop as a single phenotype increases, these cells then begin to produce inhibitory signals that force the remaining cells to develop with alternate fates.
A second area of interest is the mechanisms that control overall cellular pattern within the cochlea. Hair cells and supporting cells within the organ of Corti are arranged in precise rows that extend along the entire length of the cochlear spiral. How specific cell types become arranged into these rows is not clear, but ongoing research combines live imaging with mutant mouse models to determine how these patterns arise.
Finally, recent work in the laboratory has examined the factors that regulate the formation of tonotopic organization within the auditory systems of birds and mammals. In both birds and mammals, complex sounds are perceived through separation of those sounds into component frequencies that stimulate hair cells at different positions along the long axis of the auditory organ. To achieve this level of organization, developing hair cells and supporting cells must be able to determine their position along the tonotopic axis and to develop with frequency-specific phenotypes that are consistent for that location. Using a combination of RNA sequencing and cell biology, we are in the process of identifying the factors that regulate these processes.
Lab staff as of July 2018. Front row L-R: Tara Balasubramanian, Elizabeth Driver, Helen Maunsell, Likhitha Kolla. Second row L-R: Joey Mays, Matt Kelley, Kelly Kim. Third row L-R: Alejandro Anaya, Tessa Sanders. Back row L-R: Weise Change, Beatrice Mao, Madison Mehlferber.

